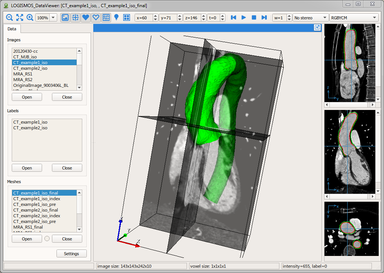
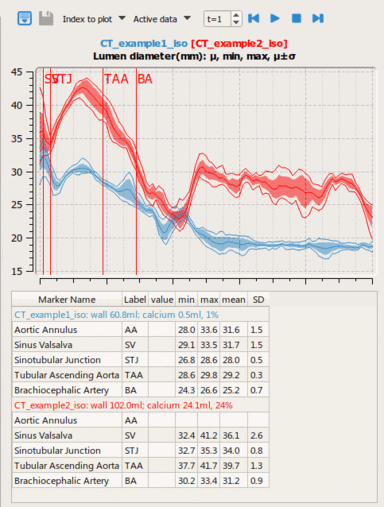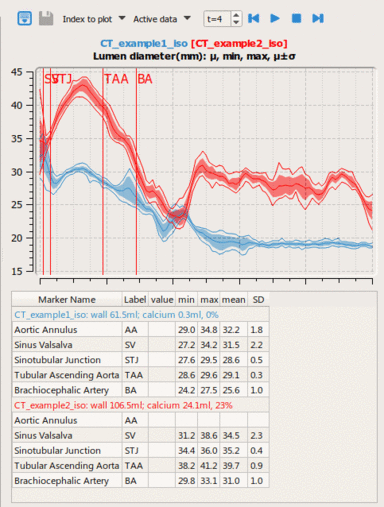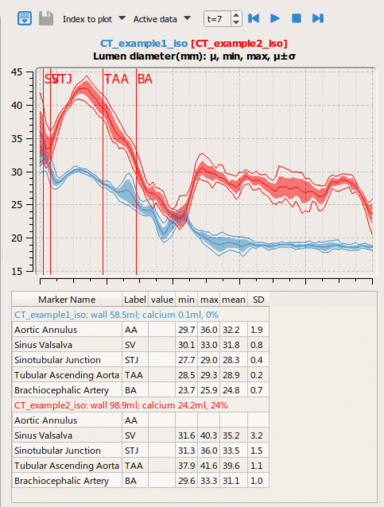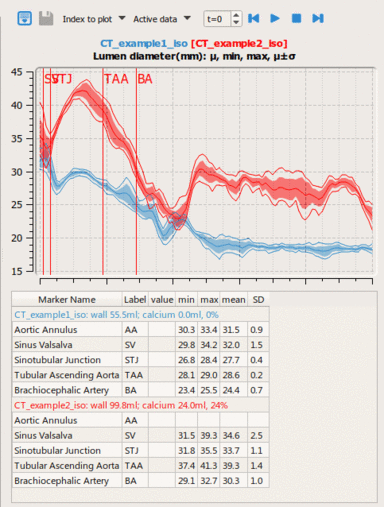
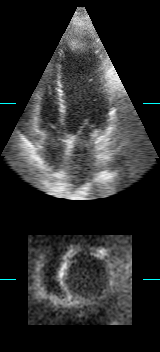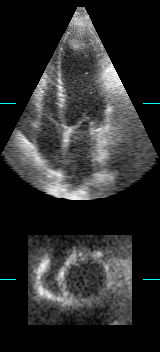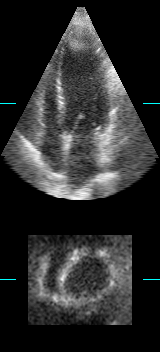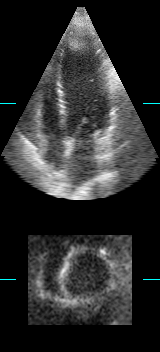
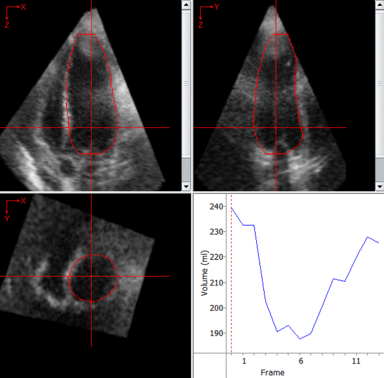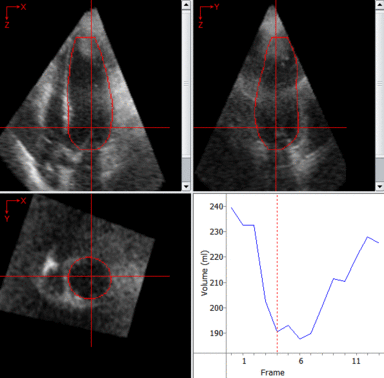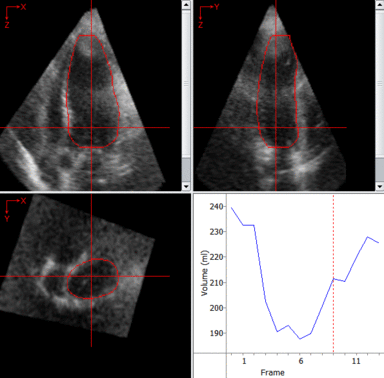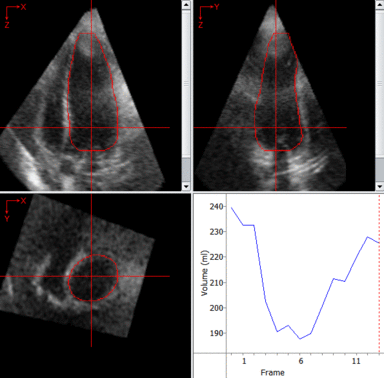
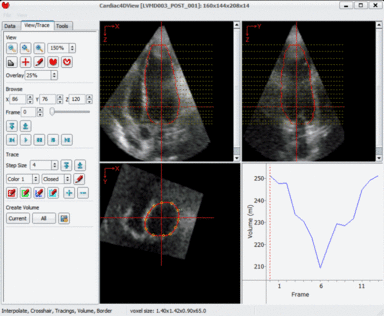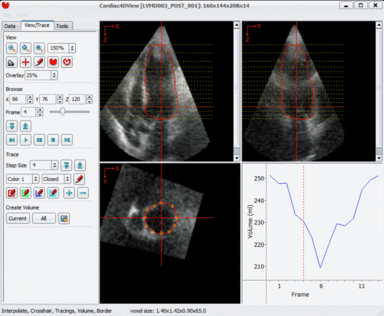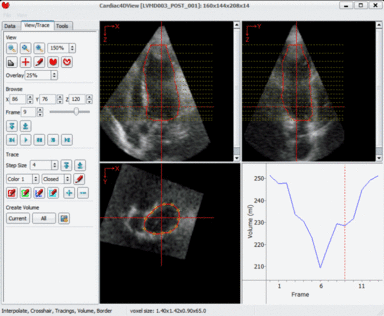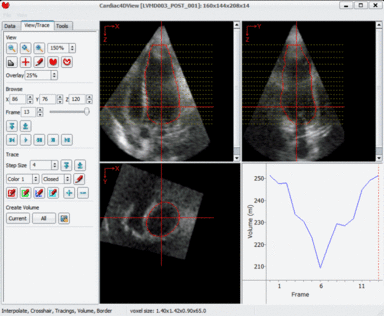
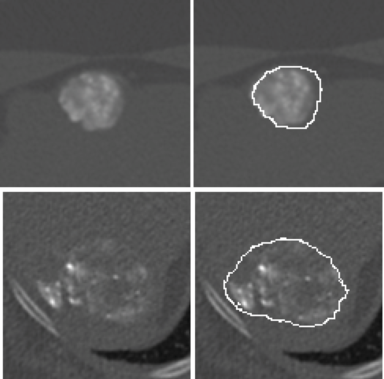
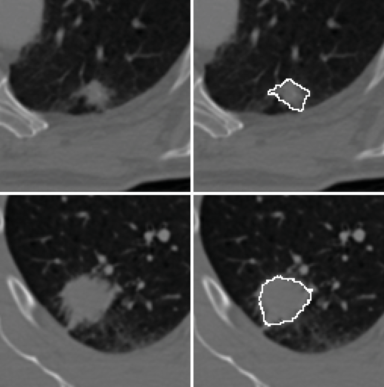
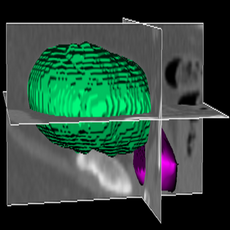
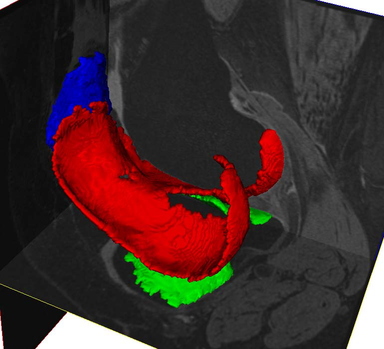
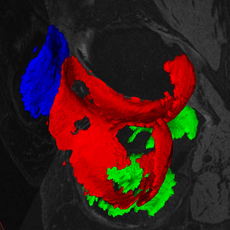
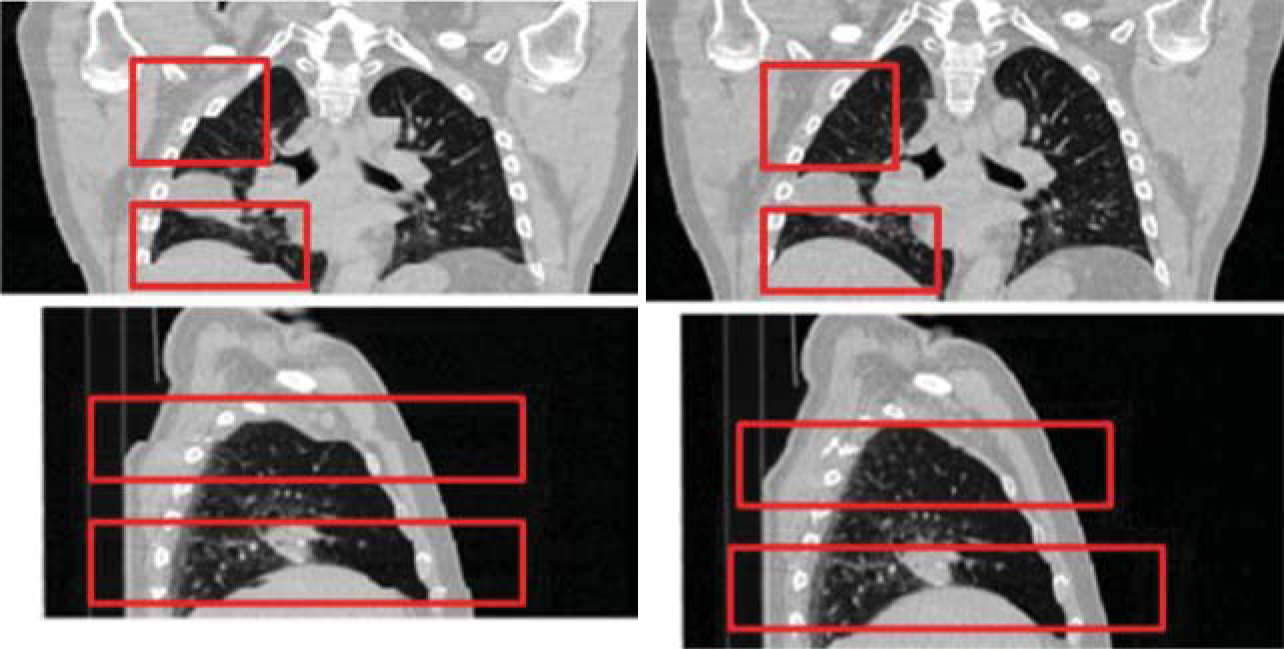
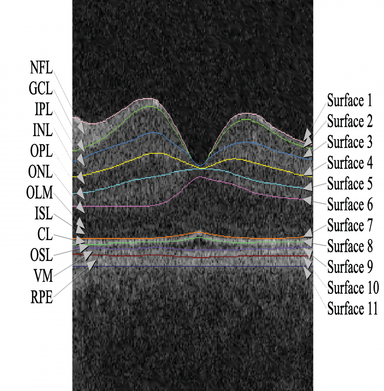
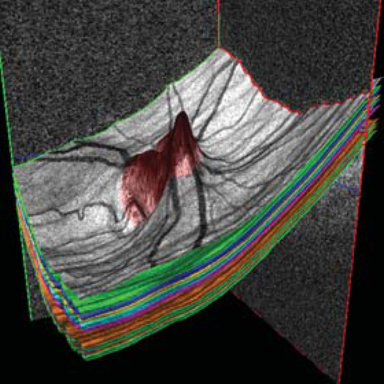
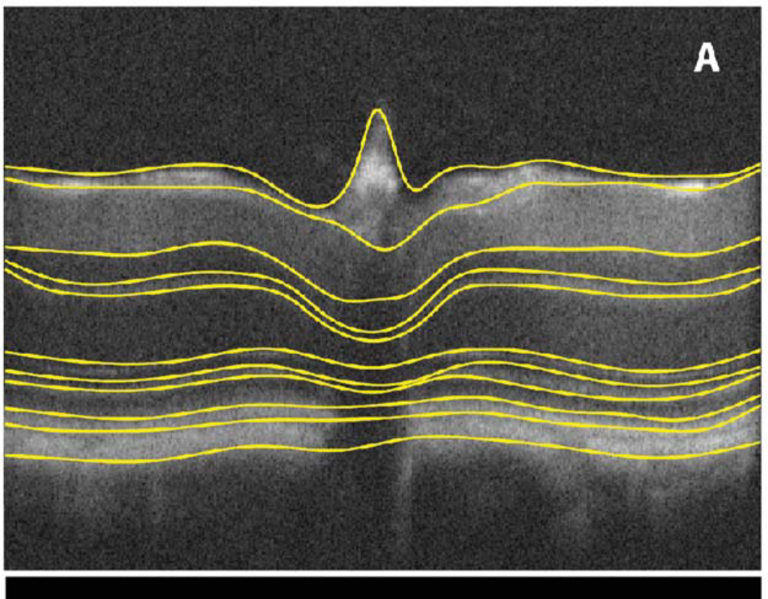
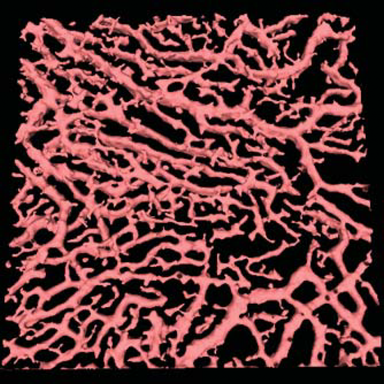
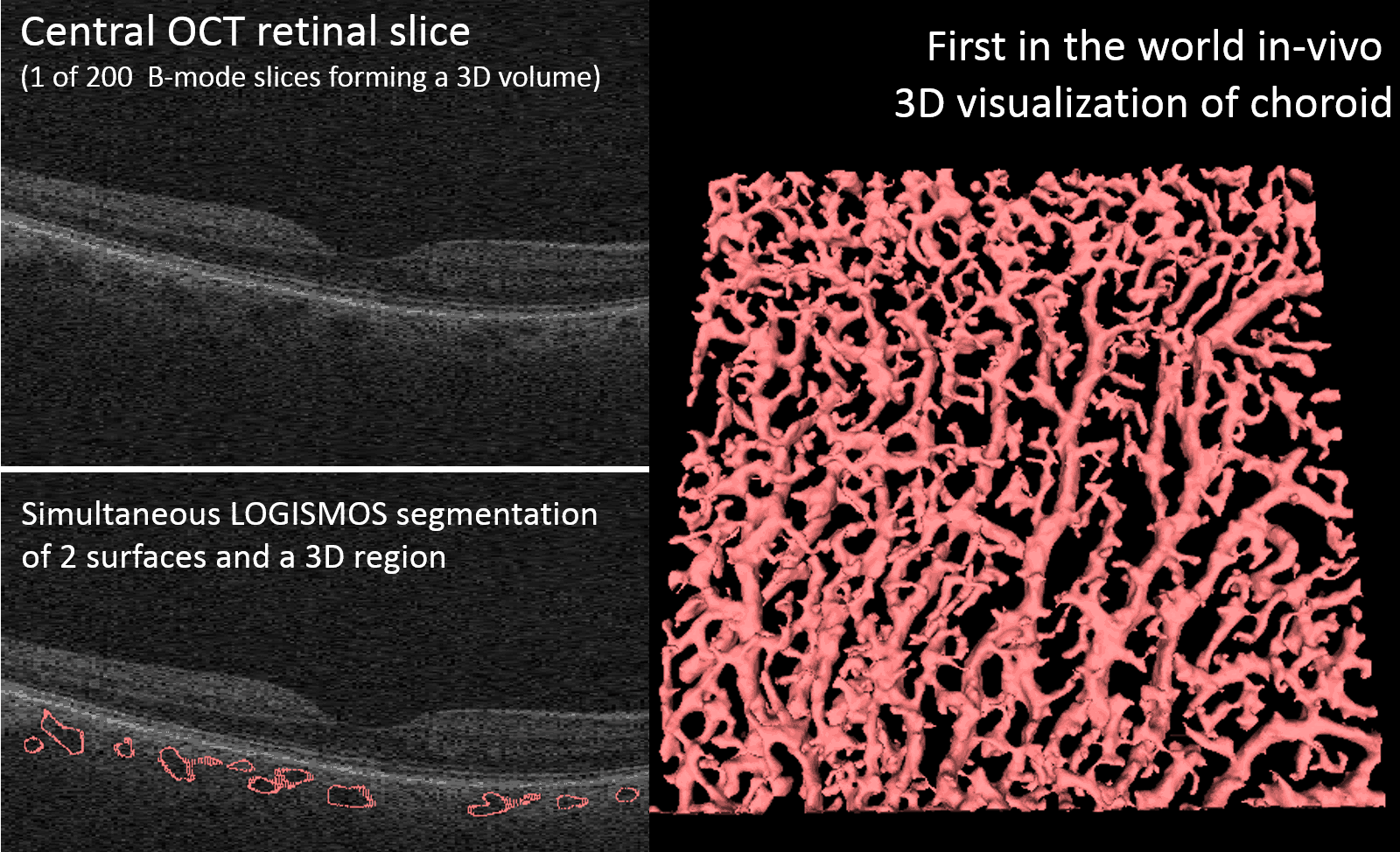
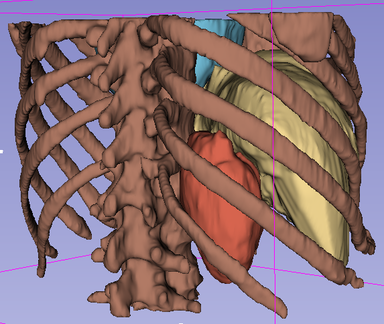
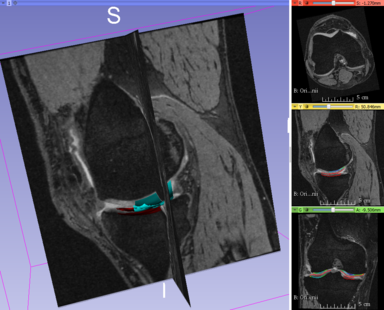
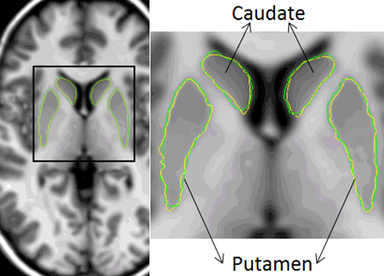
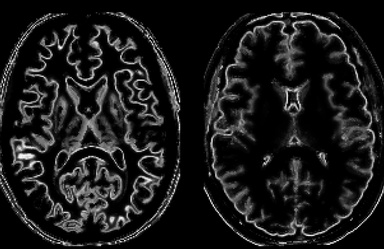
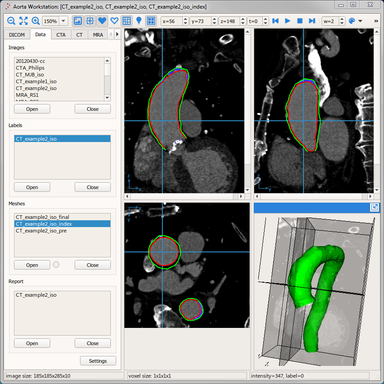
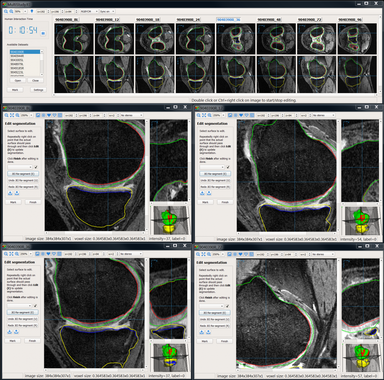
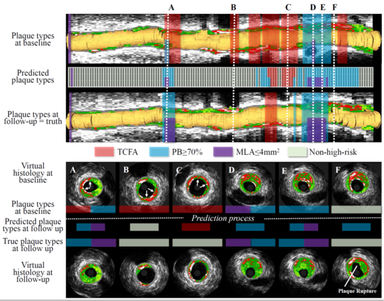
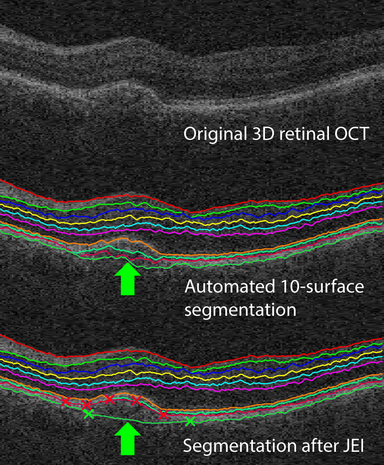
LOGISMOS segmentation framework (Layered Optimal Graph Image Segmentation for Multiple Objects and Surfaces) facilitates highly efficient multi-dimensional, multi-layered, and multi-object optimum graph-based segmentation and surface editing on image data from various modalities (CT, MR, Ultrasound, OCT, etc.).
LOGISMOS - General approach to 3D, 4D, n-D image segmentation
Sonka Team

Milan Sonka
IIBI Co-Director
The LOGISMOS framework can be applied to n-D image data and all its variants share the same general strategy:
- Pre-segmentation that yields an approximate segmentation of the objects of interest and gives information about the topological structures of the target objects;
- Mesh generation used to specify the structure of a graph GB, called base graph that defines the neighboring relations among voxels on the sought (optimal) surfaces;
- Graph construction yielding a weighted directed graph G in which each graph column corresponds to a list of vertices in G. The vertex costs of G can encode edge-based, region-based, and shape-based cost functions. Graph topology allows encoding of multi-surface/multi-object interactions;
- Graph search yields optimal surfaces corresponding to structures of interest. The sought optimal surfaces are obtained by searching for an optimal closed set in G using efficient maximum flow graph-theory algorithms;
- Highly efficient “Just-Enough Interaction” strategy for correction of local/regional mis-segmentations as/if needed.
- Application areas:
- Cardiology
- Ophthalmology
- Orthopaedics
- Neuroscience
- Pulmonology
- Radiation oncology
- Surgery
- Publications
- Code, library, application sharing
- Multi-layer 3D human macular and ONH OCT LOGISMOS+JEI segmentation entitled Iowa Reference Algorithms and murine macular segmentation programs are available as downloads for research use at www.iibi.uiowa.edu/downloads, the name Iowa Reference Algorithm is used since it allows analysis of OCT images from scanners of five tested manufacturers.
- Source code of Alpha Path-Moves is shared at github.com/sydbarrett/AlphaPathMoves
- Developed with partial support from U01 CA140206 by R. Beichel's group, cancer-related PET image analysis modules (code, libraries, apps) for the popular 3D Slicer environment are shared at github.com/QIICR/PETTumorSegmentation. Full source code is publicly available, is free for use, and shows how to implement a graph-based segmentation method with JEI functionality.
-
Support:
-
NIH-NIBIB grant EB004640 - Methodologic and algorithm development
-
- Results of the LOGISMOS project were and are used in 12 NIH (R01 EY018853, R01 EY019112, R01 HL111453, R01 CA166703, R01 NS094456, R01 EY023279, U01 CA140206, U10 EY017281, U24 CA180918, R21 AR054015, R42 HL108469, K25 CA123112), 5 Veterans Administration (1IK2RX000728, C6810-C, I01 RX001786, CENC0056P, VA-Rehab-R&D-Iowa), and 5 international research projects (Austrian Christian Doppler Soc. project OPTIMA; Ministry of Health of the Czech Republic (16-27465A, 16-28525A, 17-28784A and IKEM (MH-CZ-DRO-IKEMIN 00023001); as well as in FDA-labeled products (VIDA Diagnostics; Medical Imaging Applications LLC).
Broad Applicability of LOGISMOS n-D Segmentation Methods
Advancement of LOGISMOS Flexibility
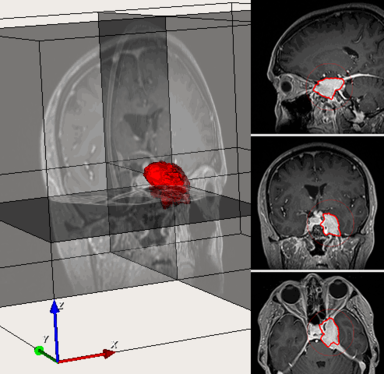
LOGISMOS+JEI in action: segment a brain tumor with simple shape, no editing required:
LOGISMOS+JEI in action: segment a brain tumor with complex shape using JEI and merging of objects:
LOGISMOS+JEI in action: segment prostate on MR T2 image:
The purpose of these videos is to demonstrate the capability of the algorithm, not to show the clinically correct segmentation.